TopHat uses Bowtie to map RNA-seq reads to a reference genome then analyzes the mapping results to identify splice junctions between exons. Improved and faster counting procedure.
Basic Analyses With Tophat Cufflinks Rnaseq Tutorial 1 Documentation
This exercise should take 1-2.
. Familiarity with Galaxy and the general concepts of RNA-seq analysis are useful for understanding this exercise. This exercise introduces these tools and guides you through a simple pipeline using some example datasets. RMATS was tested with samtools v12 and v131.
Tophat Version in GenePattern public repository. 08182016 Release of rMATS 325. 07292016 Release of rMATS 324.
While our Python installations come with many popular packages installed you may come upon a case in which you need an additional package that is not installed. 2011 TopHat is a fast splice junction mapper. Added support for multiple versions of samtools.
Optional detection of novel splice sites unannotated splice sites. For more information about the algorithm please refer to the TopHat documentation. Galaxy provides the tools necessary to creating and executing a complete RNA-seq analysis pipeline.
Aligning Rna Seq Data Ngs Analysis

Reference Based Rnaseq Data Analysis Long

Rna Seq Alignment And Visualization Youtube
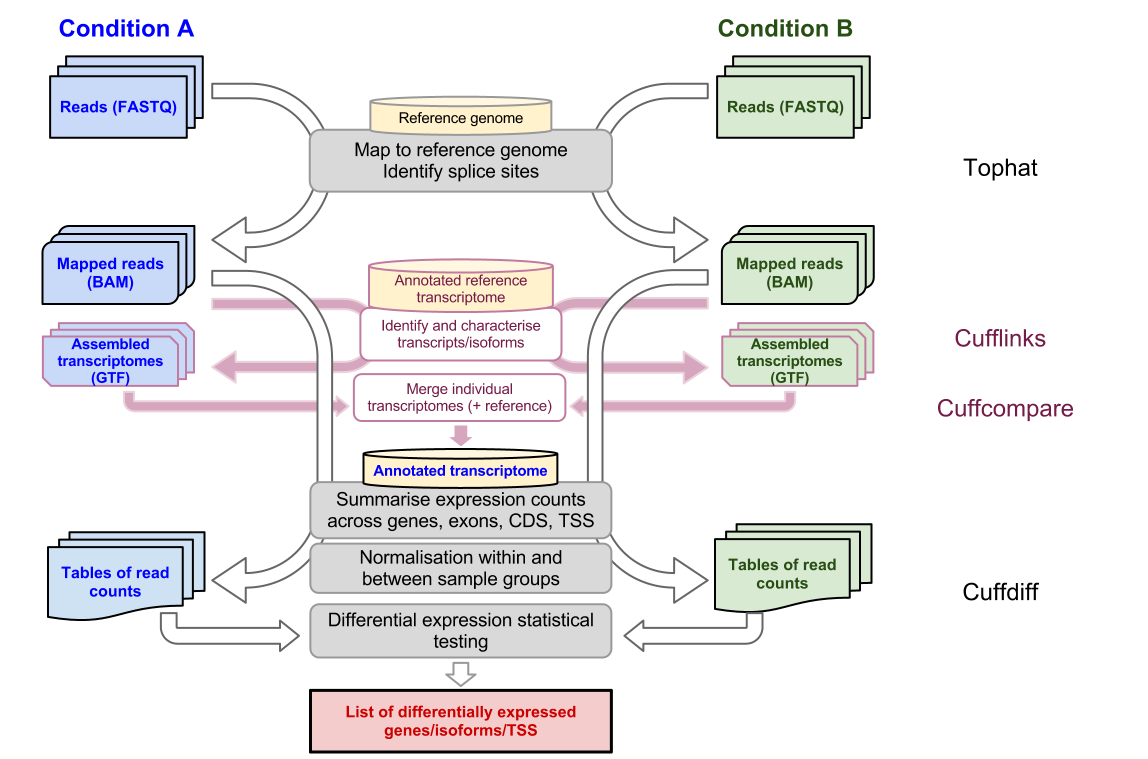
Introduction To Bulk Rnaseq Analysis Bioinformatics Documentation
The Cufflinks Rna Seq Workflow
Tophat Cufflinks Command Pipeline
Incorporating Rnaseq Tophat To Augustus Computational Biology
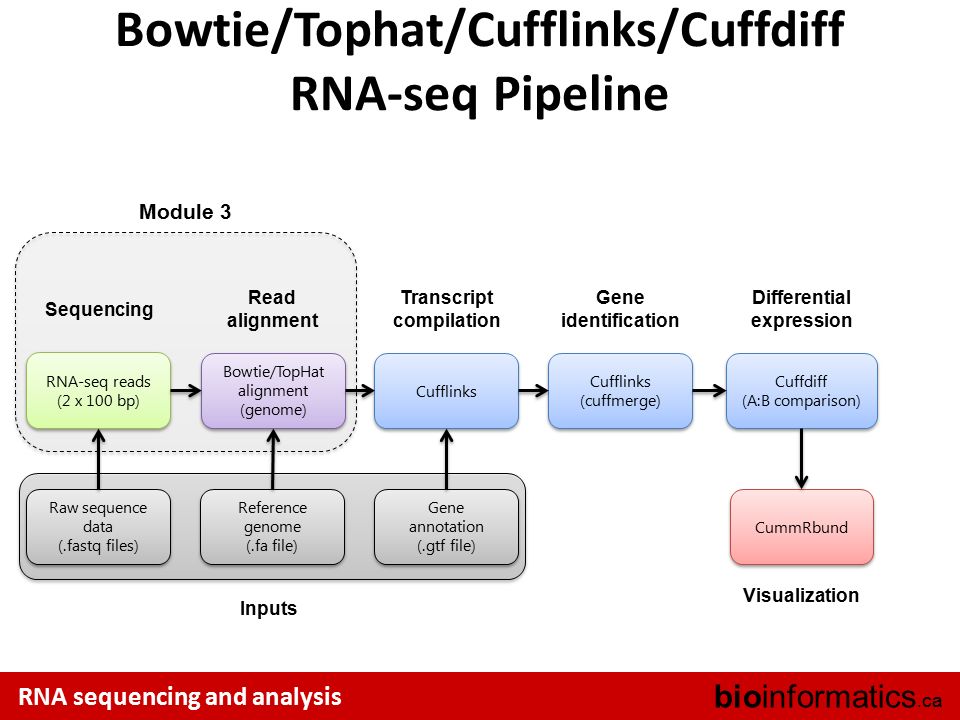
0 comments
Post a Comment